From Code to Cure: AI-Designed Proteins Arm T Cells to Precisely Target Cancer

In a breakthrough study published in Science, researchers unveiled an AI-driven platform capable of designing precision protein binders to reprogram T cells against cancer. By targeting cancer cells via peptide–MHC (pMHC) molecules, the method marks the first demonstration of fully computer-designed proteins redirecting immune responses—reducing the development timeline for novel immunotherapies from years to just a few weeks.
"We are essentially creating a new set of eyes for the immune system. Current methods for individual cancer treatment are based on finding so-called T-cell receptors in the immune system of a patient or donor that can be used for treatment. This is a very time-consuming and challenging process. Our platform designs molecular keys to target cancer cells using the AI platform, and it does so at incredible speed, so that a new lead molecule can be ready within 4-6 weeks," says Dr. Timothy P. Jenkins, Associate Professor at Technical University of Denmark (DTU) and senior author of the study.
Targeting Cancer with pMHC Precision
At the heart of this innovation lies the peptide-major histocompatibility complex (pMHC), a molecular beacon that cells use to present internal proteins to the immune system. While T cell receptors (TCRs) naturally recognize these pMHCs to detect diseased cells, engineering TCRs for therapeutic purposes has proven both complex and unpredictable.
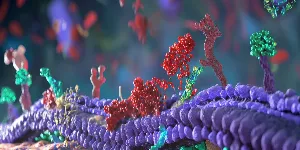
The new platform sidesteps this bottleneck by designing entirely novel “minibinders” de novo—small, synthetic proteins that mimic the function of TCRs but with greater specificity and stability. These minibinders are designed to lock onto cancer-associated pMHCs with high affinity and minimal off-target effects.
Using state-of-the-art generative modeling and molecular dynamics simulations, the team created minibinders against a well-characterized cancer antigen, NY-ESO-1 (SLLMWITQC/HLA-A*02:01), frequently expressed in melanoma and other tumors. One lead candidate exhibited nanomolar affinity, confirmed via cryo-electron microscopy, and demonstrated potent cancer cell killing when incorporated into engineered T cells.
From Algorithm to Cell Therapy
Once validated, the minibinders were fused into chimeric antigen receptors (CARs) and expressed in T cells, creating a novel therapeutic class the researchers dubbed “IMPAC-T cells.” In vitro assays showed these redesigned cells could precisely recognize and destroy NY-ESO-1–positive melanoma cells.
The platform also proved its flexibility by targeting a patient-specific neoantigen (RVTDESILSY/HLA-A*01:01) from a case of metastatic melanoma. This ability to design and validate binders against previously uncharacterized antigens opens new doors for personalized immunotherapy.
“It was incredibly exciting to take these minibinders, which were created entirely on a computer, and see them work so effectively in the laboratory,” says postdoc Kristoffer Haurum Johansen, co-author of the study and researcher at DTU.
Virtual Cross-Reactivity Screening
A crucial innovation lies in the pipeline’s in silico safety screening. Using AI models trained on large datasets of human pMHCs, the researchers filtered out binders likely to cross-react with healthy tissue, thereby reducing the risk of autoimmune toxicity before any wet-lab experiments.
“Precision in cancer treatment is crucial. By predicting and ruling out cross-reactions already in the design phase, we were able to reduce the risk associated with the designed proteins and increase the likelihood of designing a safe and effective therapy,” says Prof. Sine Reker Hadrup, a co-author of the study.
When ready, the treatment process would resemble CAR-T therapy workflows: patients provide a blood sample, immune cells are extracted and modified with AI-designed minibinders, and the reprogrammed T cells are reinfused to hunt down cancer cells.






Comments
No Comments Yet!